Knowledge Bioinformatics Lead
Knowledge Bioinformatics Lead
Biomedical data used in AI-enabled drug discovery should adhere to the FAIR Data Principles. Here I discuss why this is, how one can make data FAIR and challenges that remain.
The FAIR Data Principles — Findability, Accessibility, Interoperability and Reusability — were introduced in 2016, with the intention of improving the infrastructure that supports reusability of data, especially in machine learning applications. Despite the time that has passed since their introduction, FAIR principles are not always followed in data collection and processing. As such, many data sets we wish to use must be retrospectively made FAIR. Going forward, we must raise awareness of the issue, and ensure that everyone understands why having FAIR data is important. This blog aims to discuss the FAIR principles and why they are particularly important for biomedical data used in AI-enabled drug discovery, as well as enumerate some ways in which data can be ‘FAIRified’ and challenges that remain.
What is FAIR data?
Three of the FAIR principles — Findability, Accessibility and Reusability — are fairly straightforward to understand.
Findability refers to making both data and metadata easy to search and find by both scientists and computers. Assigning unique and persistent identifiers to data and registering or indexing it in a searchable resource are key parts of this principle.
Accessibility means that data and metadata can be retrieved by their assigned identifier using a standardised and free communications protocol (such as HTTP, FTP or SMTP). Metadata should also remain accessible, even if the data are no longer available. It’s important here to clarify that accessible data does not mean universally free to access to anyone: people wishing to access data need to meet certain criteria, such as adhering to Data Governance rules and Data Licensing requirements, to ensure that data that should be private and protected remains that way.
Reusability is related to findability but focuses on the ability of a computer or human to decide, once the data is found, if it is actually useful for the intended purpose. To make this possible, metadata should describe in detail the ‘story’ of how the data was generated. For example, what was the original purpose of generating or collecting the data, how and under what conditions was it collected or generated, how has it been processed, and what are its limitations. Usage rights and restrictions (related to accessibility) are also important to specify.
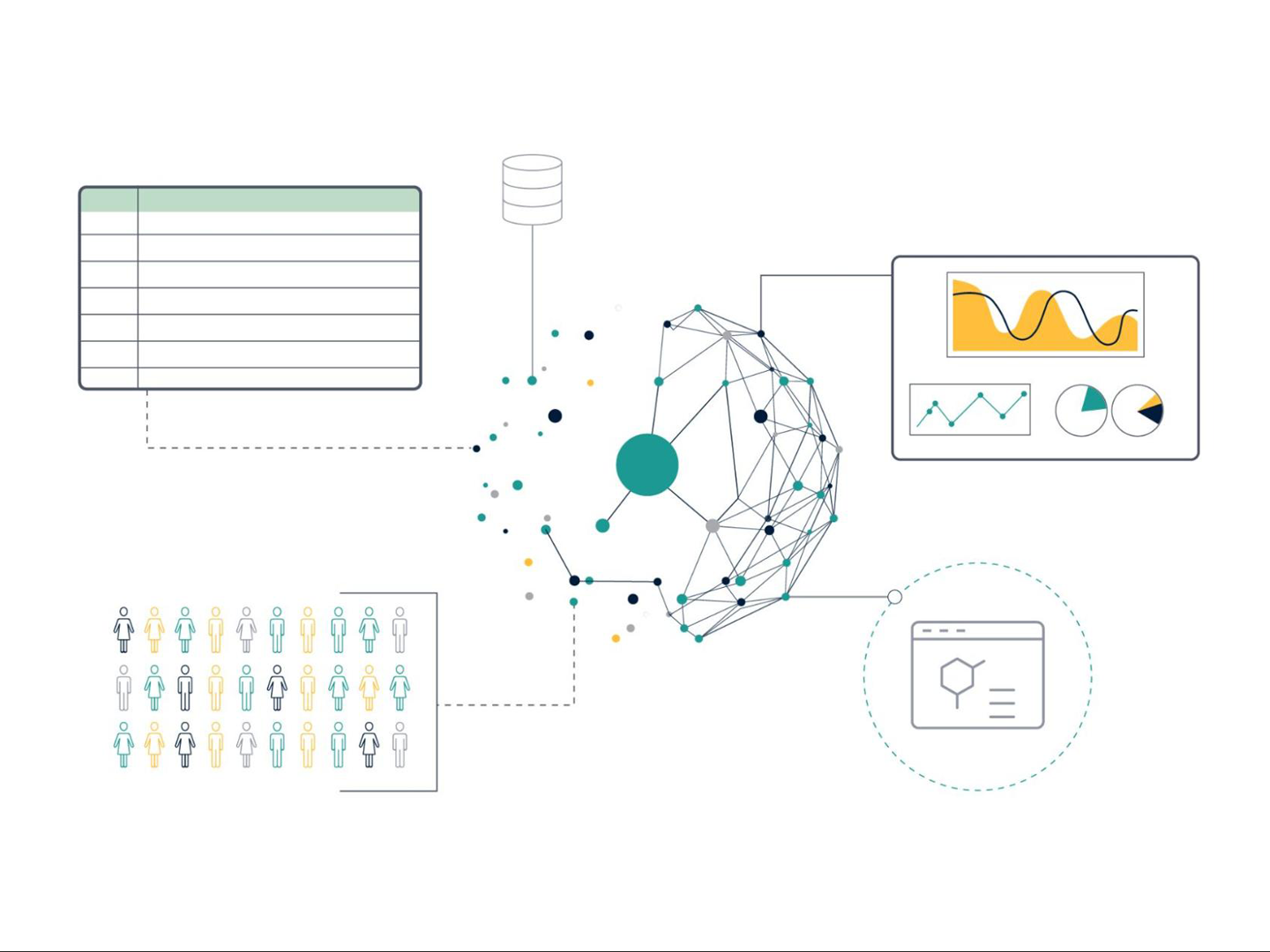
Interoperability (Figure 1) is a more difficult concept to understand and presents a true data science challenge. In essence, this means that data is formatted in such a way that facilitates linking of different data sets, allowing integration across data sources and systems. For example, it allows integration or combination of different data sources and modalities (such as structured with unstructured data or omics with chemical and biological assay data). Data that is interoperable will use a common and controlled vocabulary to describe the data (an ontology), and should specify if a data set is complete on its own or is dependent in some way on another data set, with any scientific links between the data sets clearly described.

Figure 1: Data interoperability
Why is it crucial for biomedical data to be FAIR?
Having FAIR data is important for data-driven drug discovery. Specifically, to make the best use of biomedical data in machine learning models built on knowledge graphs, such as the Benevolent PlatformTM, it is best to combine several sources and different data modalities to extract insights for one use case: for example, to generate the most insightful hypotheses for potential therapeutic targets in a given disease. However, in order to do this successfully, the data must be interoperable.
FAIR data also increases the ability of AI systems to handle large volumes of data and thereby improve model performance. This is because FAIR data is important for not only integrating data across modalities but also within modalities (such as combining many different genomics data sets) where combining different data sources allows us to increase the number of data points available. Training machine learning models using this increased amount of data will improve the performance of the models.
In addition to improving our ability to integrate publicly available data as well as licensed data sources, FAIR data can also make it more efficient to access and use data generated across departments within one pharma/biotech company or institute. This helps to remove data silos, providing more data with which we can train machine learning models, and as a side benefit, should improve data management and governance.
All data — even that which is internally generated — can’t necessarily be easily processed into a system automatically; it takes a lot of work and understanding of the data sets to integrate non-FAIR data in a way that is accurate. Therefore, adhering to the FAIR principles up front at the point of data collection should enable easier data integration and faster development of machine learning models leveraging new data.
How do we make data FAIR?
If faced with unFAIR data, what methods can be used to make it FAIR? It is beyond the scope of this short blog post to explain this in detail, but a great source of information online is the FAIR Cookbook, which provides ‘recipes’ for making life sciences data FAIR.
Ontology management and standardisation is an important part of this problem. In biomedical science, different words or phrases are often used to mean the same thing, which is challenging for making data interoperable and reusable. For example in patient records, one doctor might record a patient as having colon cancer, whereas another might record the same disease as colorectal cancer, colorectal carcinoma or bowel cancer. Sometimes patient records also record disease with different levels of specificity within a hierarchy, such as using heart failure as opposed to congestive heart failure, or left ventricular failure. As a good start on this problem, many organisations that create and manage electronic health records (including the UK Biobank and UK National Health Service) use the International Classification of Diseases (ICD) 10th Revision (ICD-10) codes for classifying diseases; in the above-mentioned heart failure hierarchy, heart failure is I50, and the more specific congestive heart failure and left ventricular failure are I50.0 and I50.1, respectively. Although this still requires some processing and mapping to be integrated into different systems, it is easier than working with records that are all free-text. Similarly, the Gene Ontology (GO) provides standard ways to describe a gene product in terms of molecular function, cellular component and biological process.
Another example of FAIR management in practice can be seen with handling of genetics data and multi-omics data, which include Whole Genome/Exome Sequencing (WGS, WES), RNA sequencing (RNAseq) and proteomics data sets. This also requires ontology standardisation on sample metadata (e.g. tissue, cell-type, treatment, patient ancestry, disease name and stage, assay technology type and experimental platform) so that genetics/omics data can be integrated with other types of data. But on top of this, other types of harmonisation are needed, such as standardising the data file format and schema. For GWAS summary statistics data, we need to harmonise variant representation, allele direction and genome build. We also need to standardise gene annotation in order to bring different omics data sets together. Harmonisation is a very big task and requires the expertise of both bioinformaticians and engineers. At BenevolentAI, we have established a squad called GGE (Genetics, Genomics, Experimental data), who are responsible for creating pipelines to harmonise all this data and making it part of our data foundations. The GGE squad also makes sure that the data quality is good, and performs testing to ensure that there is no duplication of records or missing data, and that controlled vocabulary is used.
The challenges and how they might be overcome
One major challenge is the lack of standard rules across the biomedical research industry. For example, given the complexity of the FAIRification process, curating and adding metadata to a data set may require third party involvement. This adds significant time and additional complexity to the process and delays data use. In addition, this could possibly lead to a duplication of efforts if multiple companies or groups are having the same data sets curated for their own use, resulting in wasted time and money in the drug discovery process.
As long as much of the available data is not collected according to the FAIR principles, we also must identify and prioritise the data set to be FAIRified given finite resources. This is challenging as it requires us to predict how important a data set is likely to be: will the impact of that data on our models be big enough to justify the work required to make it FAIR and enable integration in our systems, or can we find a better alternative data set elsewhere that requires less processing or is more FAIR?
There are also more technical challenges. For example, we must develop better tools to handle the more irregular ‘edge cases’. In harmonising GWAS and omics data, although the majority of entries can be made FAIR by a single standardised script, there are always a few so-called edge cases that are missing certain data fields or that require more calculations to make them FAIR. The challenge is to build a model for use in a data FAIRification pipeline that will be able to learn from the edge cases, so that the system can automatically process similar ones in the future.
It is a continual challenge to develop and improve processes to facilitate FAIRification of unFAIR data. Data management processes, policies and guidance within an organisation are needed as well to encourage data creation and collection to be FAIR from the outset. In this way, the process of increasing the amount of biomedical data that adheres to FAIR principles is both a technical and an organisational challenge.
Building The Data Foundations To Accelerate Drug Discovery
Mark Davies, BenevolentAI’s SVP Informatics and Data, discusses innovations that enable BenevolentAI to leverage biomedical data in drug discovery.
Back to blog post and videos